Mobile genetic elements in eukaryotes
Another example of transposon silencing involves plants in the genus Arabidopsis. Researchers who study these plants have found they contain more than 20 different mutator transposon sequences a type of transposon identified in maize. In wild-type plants, these sequences are methylated , or silenced.
However, in plants that are defective for one of the enzymes responsible for methylation, these transposons are transcribed. Moreover, several different mutant phenotypes have been explored in the methylation-deficient plants, and these phenotypes have been linked to transposon insertions Miura et al. Based on studies such as these, scientists know that some TEs are epigenetically silenced; in recent years, however, researchers have begun to wonder whether certain TEs might themselves have a role in epigenetic silencing.
It has taken decades for scientists to collect enough evidence to consider that maybe McClintock's speculation had a kernel of truth to it. RNAi is a naturally occurring mechanism that eukaryotes often use to regulate gene expression. Yang and Kazazian demonstrated that this results in homologous sequences that can hybridize, thereby forming a double-stranded RNA molecule that can serve as a substrate for RNAi.
Furthermore, when the investigators inhibited endogenous siRNA silencing mechanisms, they saw an increase in L1 transcripts, suggesting that transcription from L1 is indeed inhibited by siRNA. The fact that transposable elements do not always excise perfectly and can take genomic sequences along for the ride has also resulted in a phenomenon scientists call exon shuffling. Exon shuffling results in the juxtaposition of two previously unrelated exons, usually by transposition, thereby potentially creating novel gene products Moran et al.
Mobile genetic elements
The ability of transposons to increase genetic diversity, together with the ability of the genome to inhibit most TE activity, results in a balance that makes transposable elements an important part of evolution and gene regulation in all organisms that carry these sequences. Feschotte, C. Plant transposable elements: Where genetics meets genomics.
Nature Reviews Genetics 3 , — link to article. Kazazian, H. Mobile elements: Drivers of genome evolution. Science , — doi The impact of L1 retrotransposons on the human genome. Nature Genetics 19 , 19—24 link to article. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature , — link to article. Koga, A.
Vertebrate DNA transposon as a natural mutator: The medaka fish Tol2 element contributes to genetic variation without recognizable traces. Molecular Biology and Evolution 23 , — doi McLean, P. McClintock, B. Mutable loci in maize. Carnegie Institution of Washington Yearbook 50 , — link to article. Miki, Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in colon cancer. Cancer Research 52 , — Miura, A. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis.
Moran, J. Exon shuffling by L1 retrotransposition. SanMiguel, P.
Mini Review ARTICLE
Nested retrotransposons in the intergenic regions of the maize genome. Slotkin, R. Transposable elements and the epigenetic regulation of the genome.
Nature Reviews Genetics 8 , — link to article. Yang, N. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells.
Nature Structural and Molecular Biology 13 , — link to article. Restriction Enzymes. Genetic Mutation.
Functions and Utility of Alu Jumping Genes. Transposons: The Jumping Genes. DNA Transcription. What is a Gene? Colinearity and Transcription Units. Copy Number Variation.
- toshiba at300 vs sony xperia.
- Abbreviations.
- get around phone verification facebook.
- minor work permit application georgia!
Copy Number Variation and Genetic Disease. Copy Number Variation and Human Disease. Tandem Repeats and Morphological Variation. Chemical Structure of RNA. Eukaryotic Genome Complexity. RNA Functions. Pray, Ph. Transposable elements, or "jumping genes", were first identified by Barbara McClintock more than 50 years ago. Why are transposons so common in eukaryotes, and exactly what do they do? Aa Aa Aa. Transposable elements TEs , also known as "jumping genes ," are DNA sequences that move from one location on the genome to another.
Biologists were initially skeptical of McClintock's discovery. Over the next several decades, however, it became apparent that not only do TEs "jump," but they are also found in almost all organisms both prokaryotes and eukaryotes and typically in large numbers. Types of Transposons. This graph shows the contribution of DNA transposons and retrotransposons in percentage relative to the total number of transposable elements in each species.
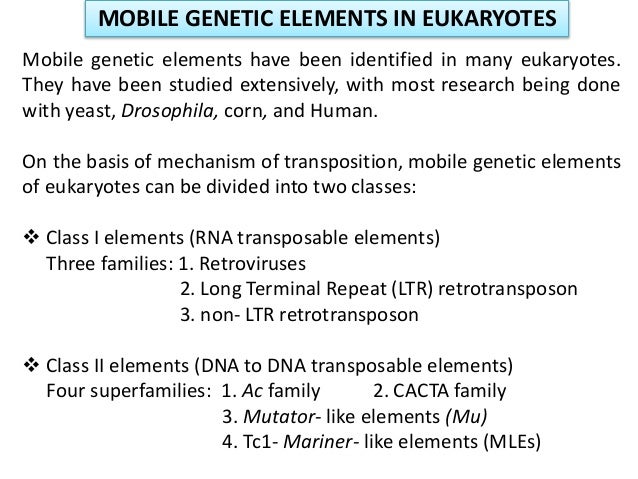
DNA Transposons. Unlike class 2 elements, class 1 elements—also known as retrotransposons—move through the action of RNA intermediaries. In other words, class 1 TEs do not encode transposase; rather, they produce RNA transcripts and then rely upon reverse transcriptase enzymes to reverse transcribe the RNA sequences back into DNA, which is then inserted into the target site. Autonomous and Nonautonomous Transposons. Both class 1 and class 2 TEs can be either autonomous or nonautonomous. Autonomous TEs can move on their own, while nonautonomous elements require the presence of other TEs in order to move.
This is because nonautonomous elements lack the gene for the transposase or reverse transcriptase that is needed for their transposition , so they must "borrow" these proteins from another element in order to move. Ac elements, for example, are autonomous because they can move on their own, whereas Ds elements are nonautonomous because they require the presence of Ac in order to transpose. Silencing and Transposons. As opposed to L1 , most TEs appear to be silent—in other words, these elements do not produce a phenotypic effect, nor do they actively move around the genome.
At least that has been the general scientific consensus. Some silenced TEs are inactive because they have mutations that affect their ability to move from one chromosomal location to another; others are perfectly intact and capable of moving but are kept inactive by epigenetic defense mechanisms such as DNA methylation , chromatin remodeling , and miRNAs. In chromatin remodeling , for example, chemical modifications to the chromatin proteins cause chromatin to become so constricted in certain areas of the genome that the genes and TEs in those areas are silenced because transcription enzymes simply cannot access them.
Because transposon movement can be destructive, it is not surprising that most of the transposon sequences in the human genome are silent, thus allowing this genome to remain relatively stable, despite the prevalence of TEs. Moreover, research suggests that even these few remaining active transposons are inhibited from jumping in a variety of ways that go beyond epigenetic silencing.
DNA Transposons and the Evolution of Eukaryotic Genomes | Annual Review of Genetics
Transposons Are Not Always Destructive. Not all transposon jumping results in deleterious effects. In fact, transposons can drive the evolution of genomes by facilitating the translocation of genomic sequences, the shuffling of exons, and the repair of double-stranded breaks. Insertions and transposition can also alter gene regulatory regions and phenotypes. In the case of medaka fish , for instance, the Tol2 DNA transposon is directly linked to pigmentation.
One highly inbred line of these fish was shown to have a variety of pigmentation patterns. Transposable elements that utilize reverse transcriptase to transpose through an RNA intermediate are termed retrotransposons. They are widespread among eukaryotes and are generally divided into two classes.
Viral retrotransposons have properties similar to those of retroviruses. For instance, they display long terminal repeats, or LTRs, as shown in Figure Two examples of viral retrotransposons are the yeast Ty elements, and the Drosophila copia elements. A schematic representation of a viral retrotransposon. Such elements have two LTRs long terminal repeats that flank a central region that encodes specific protein functions. From H. Lodish, D. Baltimore, A. Berk, S. Zipursky, P.